Highlighted Products
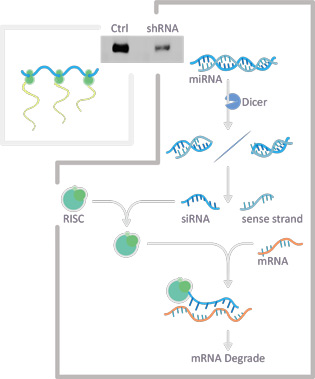 RNA interference (RNAi) refers to a cellular mechanism of post-transcriptional gene regulation through the actions of non-coding microRNAs (miRNAs). These miRNAs are generated from a longer pri-microRNA transcribed from genomic DNA. Processing of this pri-microRNA occurs through the nuclear microprocessor complex and then by the cytoplasmic Dicer complex to produce a double-stranded RNA duplex. One of the strands is then taken up into the RISC complex, where the sequence of the strand will guide the complex to a specific mRNA for degradation or inactivation.
RNA interference (RNAi) refers to a cellular mechanism of post-transcriptional gene regulation through the actions of non-coding microRNAs (miRNAs). These miRNAs are generated from a longer pri-microRNA transcribed from genomic DNA. Processing of this pri-microRNA occurs through the nuclear microprocessor complex and then by the cytoplasmic Dicer complex to produce a double-stranded RNA duplex. One of the strands is then taken up into the RISC complex, where the sequence of the strand will guide the complex to a specific mRNA for degradation or inactivation.
Scientists realized that they could use this system to diminish or even temporarily eradicate the cellular levels of virtually any transcript or protein. They could do this by synthesizing very short (19-21 nucleotides) RNA duplexes that resemble the products fashioned by the microprocessor and Dicer complexes. These small interfering RNAs (siRNAs) are transfected into cells where they associate with RISC and subsequently bind to their mRNA targets through complete complementarity of the sequences of the siRNA and the mRNA. The RISC complex then degrades/inactivates the mRNA before it can be translated, thereby leading to decreased levels of the corresponding protein.
The challenges with RNA interference technology
One challenge with siRNAs is that they are subject to degradation themselves, with only about 1% of the transfected duplex remaining two days after introduction. This means that large amounts must be transfected in order to maximize the chance of success. In addition, knockdown of the specific protein is transient. One way to address these two issues is to introduce into cells plasmids expressing short hairpin RNAs (shRNAs) that are converted into siRNAs by the endogenous RNAi machinery. This approach allows the persistent expression of siRNAs within the cell, thereby facilitating long-term knockdown of the target transcript. Various inducible or otherwise promoter-specific vectors can be used to create even more refined control of shRNA expression.
GeneTex engages RNA interference technology to validate antibody specificity
At GeneTex, we use an shRNA-mediated RNAi protocol employing a lentiviral expression plasmid optimized for the expression of shRNAs. Once transported into the nucleus, the plasmid directs transcription of the shRNA that is subsequently processed like the miRNAs described above. This makes shRNA more efficient. In addition, the knockdown is more stable over time as the shRNA is constantly expressed. Interestingly, only a few copies of the shRNA-expressing vector need to be integrated into the cell's genome in order to maintain knockdown. This decreases the risk of off-target effects, which is a more prominent problem with transfected siRNAs at their higher introduced concentrations.
GeneTex complements these efforts with CRISPR/Cas9-mediated KO cell lines
GeneTex will continue to expand its utilization of shRNA as a key component of its antibody validation program. In addition, we will incorporate other methodologies to guarantee the specificity of our products, including application of CRISPR/Cas9 technology. Made possible by our collaborative relationship with the genome editing technology company EdiGene, GeneTex will use lysates from EdiGene’s knockout (KO) cell lines to definitively demonstrate antibody specificity. At GeneTex, we understand the importance of quality reagents to your research. We are committed to providing best-in-class reagents and leading the research antibody industry in antibody validation.
 RNA interference (RNAi) refers to a cellular mechanism of post-transcriptional gene regulation through the actions of non-coding microRNAs (miRNAs). These miRNAs are generated from a longer pri-microRNA transcribed from genomic DNA. Processing of this pri-microRNA occurs through the nuclear microprocessor complex and then by the cytoplasmic Dicer complex to produce a double-stranded RNA duplex. One of the strands is then taken up into the RISC complex, where the sequence of the strand will guide the complex to a specific mRNA for degradation or inactivation.
RNA interference (RNAi) refers to a cellular mechanism of post-transcriptional gene regulation through the actions of non-coding microRNAs (miRNAs). These miRNAs are generated from a longer pri-microRNA transcribed from genomic DNA. Processing of this pri-microRNA occurs through the nuclear microprocessor complex and then by the cytoplasmic Dicer complex to produce a double-stranded RNA duplex. One of the strands is then taken up into the RISC complex, where the sequence of the strand will guide the complex to a specific mRNA for degradation or inactivation.